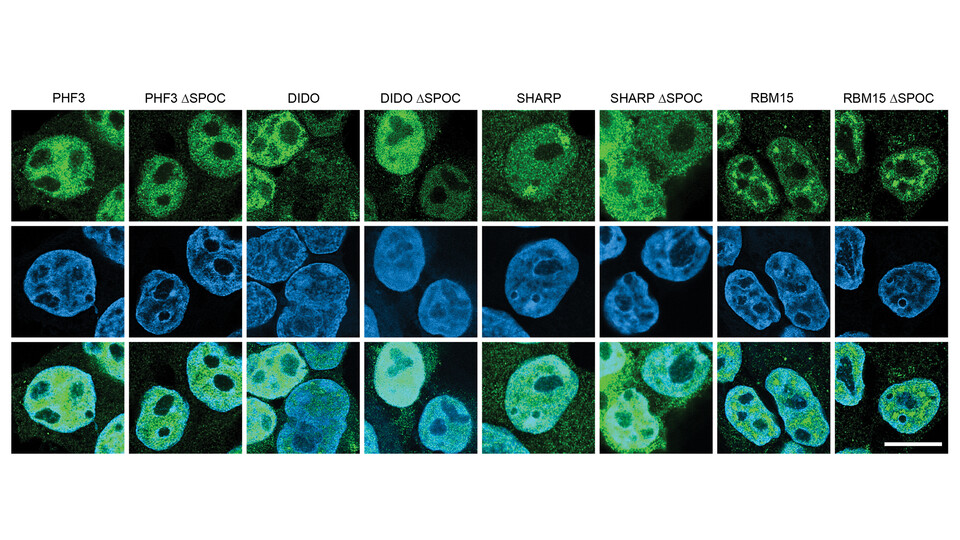

The simplest proteins are built from a single chain of amino acids that fold into an energetically stable, three-dimensional structure. Many complex proteins, however, contain multiple domains, sub-structures that can fold into a stable compact structure independent of the rest of the protein. During the course of evolution, large proteins with complex functions have been created by the assembly of domains in a combinatorial, modular manner. One such domain is called SPOC (Spen orthologue and paralogue C-terminal), which is found in six human proteins. In 2021, the lab of Dea Slade demonstrated that the SPOC domain of a protein called PHF3 recognizes phosphorylated residues in the C-terminal domain (CTD) of RNA polymerase II, thereby linking PHF3 to the regulation of gene expression. Whether CTD domain binding is unique to PHF3 or if other SPOC domains possess the same ability has, however, so far not been addressed.
The Slade lab has now teamed up with scientists from the Max Delbrück Center (Berlin) along with the groups of Kristina Djinovic Carugo and Bojan Zagrovic at the Max Perutz Labs to study the remaining five known SPOC proteins in humans. “We found that these SPOC domains also bind to the C-terminal domain of RNA polymerase II. SPOC is therefore a universal CTD reader, but SPOC domain-containing proteins exhibit different binding specificities”, explains Dea Slade. By recognizing differentially phosphorylated CTD residues, these proteins regulate transcription at different stages.
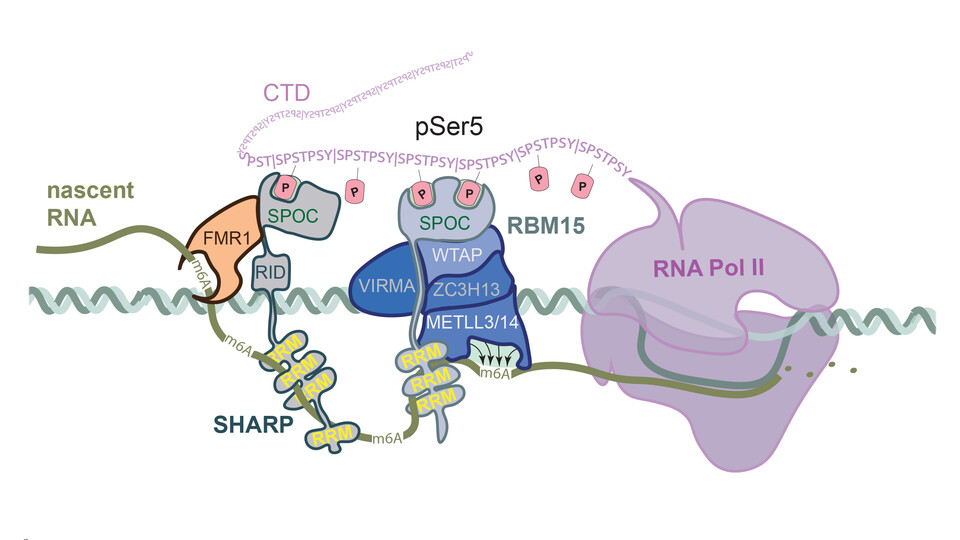
Supported by the mass spectrometry facility at the Max Perutz Labs, the team investigated potential binding partners of SPOC proteins apart from RNA polymerase II. They discovered that RBM15 and SHARP, members of the SPEN family, interact with readers and writers of N6-methyladenosine (m6A), one of the most abundant RNA modifications, which plays a key role in regulating RNA stability. “RBM15 was previously characterized as a component of the m6A complex, but it was unclear how it recruits the complex onto RNA. We show that it is the RBM15 SPOC domain that brings the whole complex to the target RNA”, says Dea Slade. In this way, transcription is directly linked to the regulation of RNA stability. However, many questions regarding the fine tuning of the interactions between SPOC proteins and their partners remain. “SPOC proteins share binding partners with similar binding affinities, raising the question of how these interactions are regulated in the cell and what the functional consequences of switching between binding partners are”, concludes Dea Slade.
Lisa-Marie Appel, Vedran Franke, Johannes Benedum, Irina Grishkovskaya, Xué Strobl, Anton Polyansky, Gregor Ammann, Sebastian Platzer, Andrea Neudolt, Anna Wunder, Lena Walch, Stefanie Kaiser, Bojan Zagrovic, Kristina Djinovic-Carugo, Altuna Akalin, Dea Slade: The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators. Nature Communications 2022
https://www.nature.com/articles/s41467-023-35853-1