
The MAP kinase cascade is a central signaling pathway that transmits extracellular signals from the cell surface to the nucleus, leading to changes in gene expression and cellular responses such as growth, differentiation, and proliferation. RAF1, a serine/threonine kinase, serves as a critical link between membrane-bound small G-proteins, such as RAS, and the cascade. However, some cellular phenotypes – such as impaired autophagy upon RAF1 ablation – have hinted at non-canonical roles for RAF1.
“We were curious whether autophagy-related proteins might be substrates of RAF1”, first author Stefanie Toifl explains. The researchers used a proximity-based mass spectrometry approach to identify 70 candidate proteins in the proxisome of RAF1 that are linked to autophagy and the lysosome. Previously, MEK1 and MEK2 were the only known RAF1 substrates in the MAP kinase pathway.
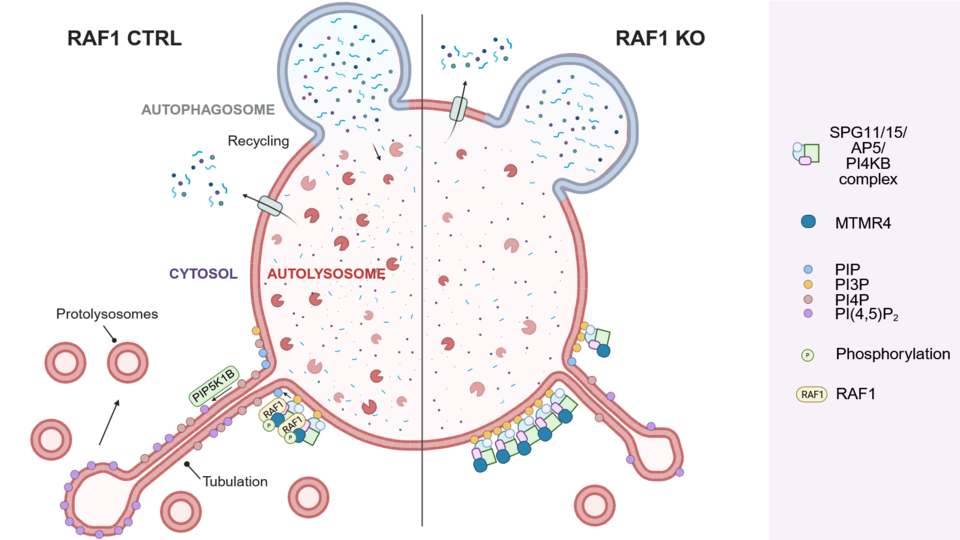
Among the identified RAF1 interaction candidates, the lipid phosphatase MTMR4 stood out: computational analysis revealed that it contains a RAF1 phosphorylation site similar to the RAF1 phosphorylation site in MEK1. Consistent with an interaction, the authors found that RAF1 and MTMR4 co-localize on lysosomes and autolysosomes. “RAF1 localization at lysosomes and autolysosomes is, in itself, a novel finding”, Stefanie says. Based on these insights, the team have proposed a model in which RAF1 phosphorylates MTMR4 at the autolysosome, thereby regulating phospholipid turnover. “RAF1’s regulation of MTMR4 is crucial for the activation and maintenance of membrane tubulation, a process essential for regenerating lysosomes”, Stefanie adds.
What excited the researchers most were the striking microscopic videos they captured while exploring the lysosomal compartment. Enhanced by super-resolution imaging, these movies afforded detailed insights. In a spin-off from the main project, the team also developed a faster method to analyze the phospholipid composition of lysosomes using a published protocol for the immunoprecipitation of lysosomes called Lyso-IP combined with a dot blot technique – a tool that could support further discoveries and will be the subject of a future publication.
DOI: 10.1016/j.celrep.2025.115490