SLAMseq resolves the kinetics of maternal and zygotic gene expression during early zebrafish embryogenesis.
2023 Cell reports;42(2):112070.
PMID: 36757845
Bhat Pooja, Cabrera-Quio Luis E, Herzog Veronika A, Fasching Nina, Pauli Andrea, Ameres Stefan L
Time-Resolved Small RNA Sequencing Unravels the Molecular Principles of MicroRNA Homeostasis.
2019 Molecular cell(4)
PMID: 31350118
Reichholf Brian, Herzog Veronika A, Fasching Nina, Manzenreither Raphael A, Sowemimo Ivica, Ameres Stefan L
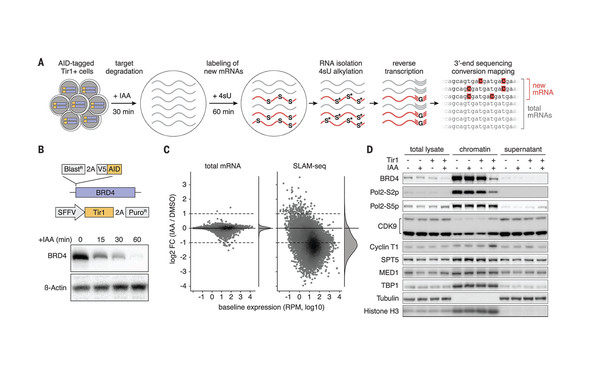
SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis.
2018 Science (New York, N.Y.)(6390)
PMID: 29622725
Muhar Matthias, Ebert Anja, Neumann Tobias, Umkehrer Christian, Jude Julian, Wieshofer Corinna, Rescheneder Philipp, Lipp Jesse J, Herzog Veronika A, Reichholf Brian, Cisneros David A, Hoffmann Thomas, Schlapansky Moritz F, Bhat Pooja, von Haeseler Arndt, Köcher Thomas, Obenauf Anna C, Popow Johannes, Ameres Stefan L, Zuber Johannes
Cell-type specific sequencing of microRNAs from complex animal tissues.
2018 Nature methods(4)
PMID: 29481550
Alberti Chiara, Manzenreither Raphael A, Sowemimo Ivica, Burkard Thomas R, Wang Jingkui, Mahofsky Katharina, Ameres Stefan L, Cochella Luisa
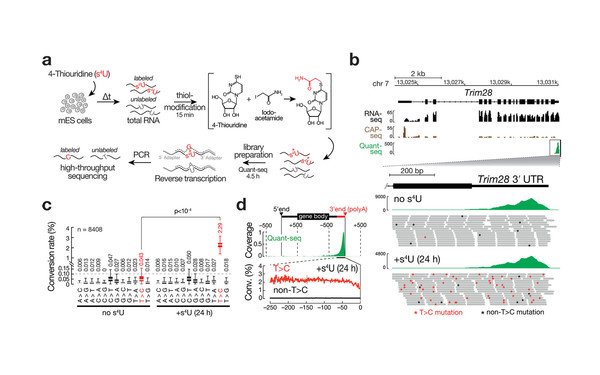
Thiol-linked alkylation of RNA to assess expression dynamics.
2017 Nature methods(12)
PMID: 28945705
Herzog Veronika A, Reichholf Brian, Neumann Tobias, Rescheneder Philipp, Bhat Pooja, Burkard Thomas R, Wlotzka Wiebke, von Haeseler Arndt, Zuber Johannes, Ameres Stefan L
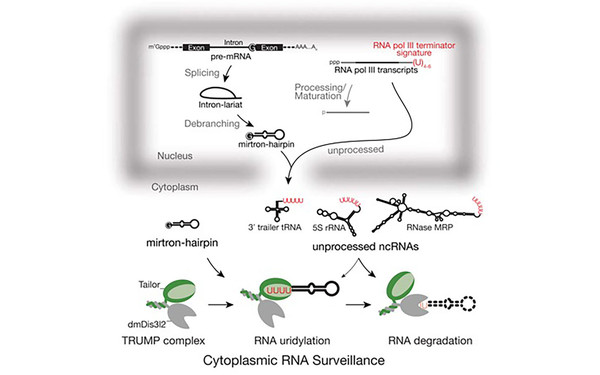
Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila.
2016 The EMBO journal(22)
PMID: 27729457
Reimão-Pinto Madalena M, Manzenreither Raphael A, Burkard Thomas R, Sledz Pawel, Jinek Martin, Mechtler Karl, Ameres Stefan L
Uridylation of RNA Hairpins by Tailor Confines the Emergence of MicroRNAs in Drosophila.
2015 Molecular cell(2)
PMID: 26145176
Reimão-Pinto Madalena M, Ignatova Valentina, Burkard Thomas R, Hung Jui-Hung, Manzenreither Raphael A, Sowemimo Ivica, Herzog Veronika A, Reichholf Brian, Fariña-Lopez Sara, Ameres Stefan L
Long-term, efficient inhibition of microRNA function in mice using rAAV vectors.
2012 Nature methods(4)
PMID: 22388288
Xie Jun, Ameres Stefan L, Friedline Randall, Hung Jui-Hung, Zhang Yu, Xie Qing, Zhong Li, Su Qin, He Ran, Li Mengxin, Li Huapeng, Mu Xin, Zhang Hongwei, Broderick Jennifer A, Kim Jason K, Weng Zhiping, Flotte Terence R, Zamore Phillip D, Gao Guangping
Target RNA-directed trimming and tailing of small silencing RNAs.
2010 Science (New York, N.Y.)(5985)
PMID: 20558712
Ameres Stefan L, Horwich Michael D, Hung Jui-Hung, Xu Jia, Ghildiyal Megha, Weng Zhiping, Zamore Phillip D
Molecular basis for target RNA recognition and cleavage by human RISC.
2007 Cell;130(1):101, 112, 101-12.
PMID: 17632058
Ameres Stefan Ludwig, Martinez Javier, Schroeder Renée