
TRIM proteins – named after their characteristic Tripartite Motif – form a large family of proteins involved in innate immunity, cellular homeostasis, and antiviral defense. Among them, TRIM52 stands out for having been lost or pseudogenized in most mammals but retained in humans and some primates. Notably, TRIM52 is upregulated in several cancer types, and its loss has been shown to reduce cellular fitness. Group leader Gijs Versteeg adds: “TRIM52 is one of the most unstable proteins in the cell, turning over every four minutes, but its precise role is currently unknown. This raises the question: Why does a cell invest so much energy to produce a protein, only to degrade it almost immediately?”
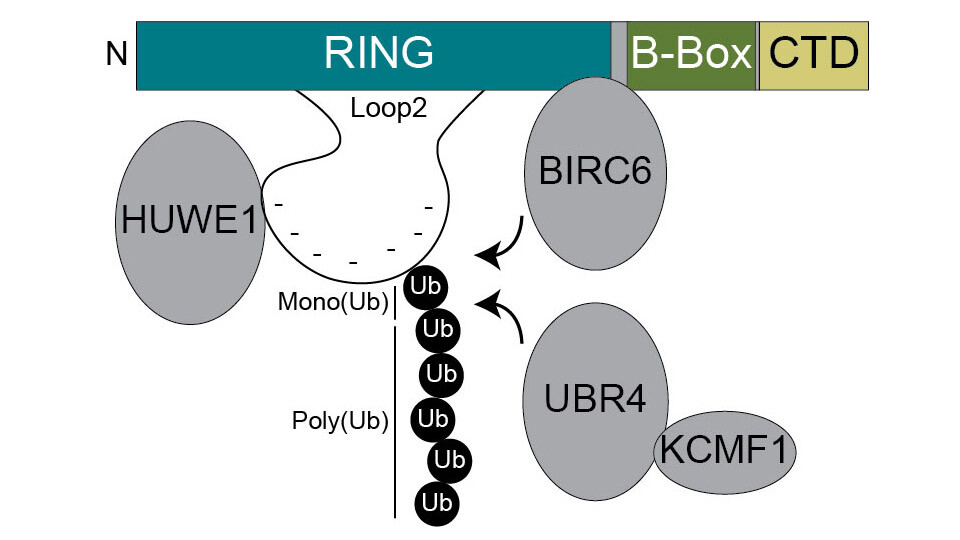
Seeking answers to this fundamental question, the Versteeg lab focused on identifying the factors responsible for the degradation of TRIM52. Although TRIM52 contains a RING finger domain – a common site for ubiquitination – the domain is extended, raising the possibility that this feature contributes to its rapid turnover. “We discovered that several giant E3 ligases, responsible for marking proteins for degradation, bind specifically to the extended loop 2 within TRIM52’s RING finger”, Gijs says.
The rapid turnover of TRIM52, however, remained puzzling. Previous observations made by the lab suggested that TRIM52 loss affects the activity of p53, a key regulator of the cell cycle and well-known tumor suppressor. To explore this further, the team conducted an unbiased genetic screen in which they identified several genetic interactors linked to the non-homologous end-joining (NHEJ) and Homology-Dependent Repair (HDR) DNA-damage response pathway, indicating that TRIM52 may participate in DNA repair processes.
To better understand the specific type of DNA damage TRIM52 addresses, first author Alexandra and her colleagues employed proximity labeling. Among the strongest hits were proteins involved in resolving DNA lesions made by topoisomerase 2 – an enzyme that prevents DNA helix supercoiling by creating temporary double-strand breaks to relieve torsional stress during replication and transcription. Alexandra explains: “Our findings suggest that TRIM52 may play a crucial role in maintaining genome stability by helping cells resolve DNA double-strand breaks, particularly those caused by topoisomerase activity during normal cellular processes.”
The Versteeg lab’s findings bring them one step closer to understanding TRIM52’s biological role. “The big question for the future, however, remains why TRIM52 is turned over so rapidly in cells, and how this is connected to its cellular function”, Gijs emphasizes.
The study was carried out in collaboration with the Clausen lab at the Institute of Molecular Pathology (IMP) in Vienna and the Cochella lab at the Johns Hopkins University in Baltimore, USA.
DOI: 10.1038/s