
Interferons are frontline defenders of cells and tissues, activating the JAK/STAT signaling cascade to drive expression of immune-regulatory genes. IFNγ, in particular, promotes antibacterial responses like phagocytosis and reactive oxygen species production, and determined effectiveness of immune checkpoint inhibition. Regulation of the signaling pathway occurs on multiple levels, including RNA processing, but identifying post-transcriptional regulators has proven challenging due to their essentiality.
To overcome the lethality of gene knockouts, the authors employed an inducible CRISPR-Cas9 system in which Cas9 expression is triggered by doxycycline. By tightly controlling the timing of Cas9 expression, the researchers created a short window – a ‘hypomorphic’ state – during which loss of gene expression coupled with protein turnover reveals phenotypic effects before cells lose viability. Perutz group leader Gijs Versteeg says: “On the basis of this screen, we found that ERH and other post-transcriptional RNA regulators are critical for IFNγ signaling. Without them, this immune response just doesn’t happen.”
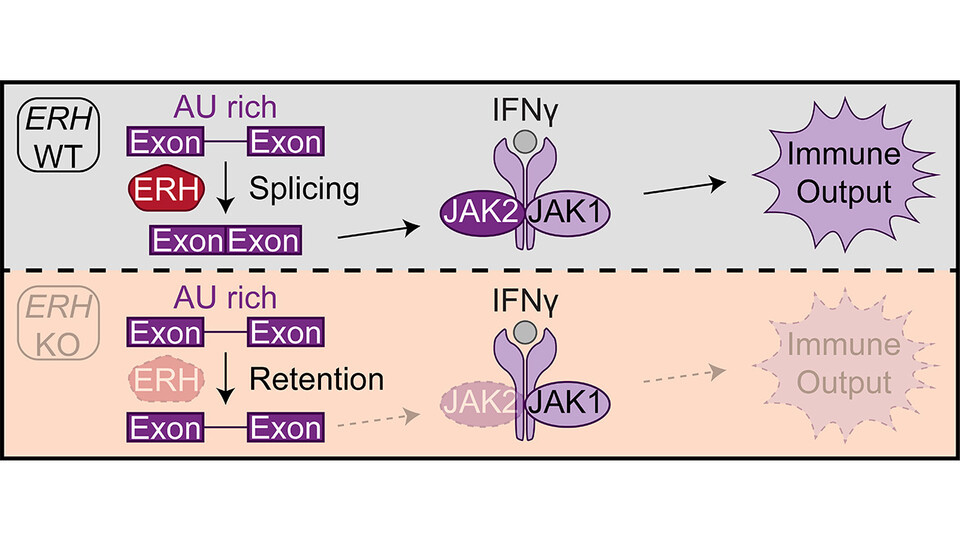
The scientists were able to demonstrate that without ERH, JAK2 transcripts are improperly processed, retained in the nucleus, and degraded, leading to a defect in immune signaling. Proper splicing and export of JAK2 transcripts are essential for an effective IFNγ response. “ERH ensures that these steps are carried out correctly, especially for mRNAs containing AU-rich introns”, says first author and PhD student Adrian Söderholm. “Since retained introns have thus far been described to be GC-rich, this suggests that cells may employ a novel, as yet uncharacterized, co- or post-transcriptional regulatory mechanism.”
Looking ahead, the team aims to understand why certain transcripts are particularly affected by the loss of RNA processing regulators like ERH. One approach involves transferring defective introns into artificial reporter RNAs to test whether the defect is sequence intrinsic; another explores whether nuclear positioning of transcripts affects ERH-dependent splicing. The study was conducted in collaboration with researchers at the Perutz, the Institute of Molecular Pathology (IMP) in Vienna, the Institute of Molecular Biology in Mainz, and the Institute of Molecular Biosciences in Frankfurt, Germany.
DOI: 10.1093/nar/gkaf545