The Question
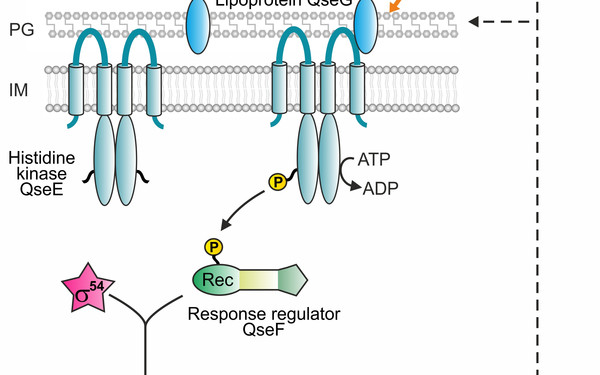
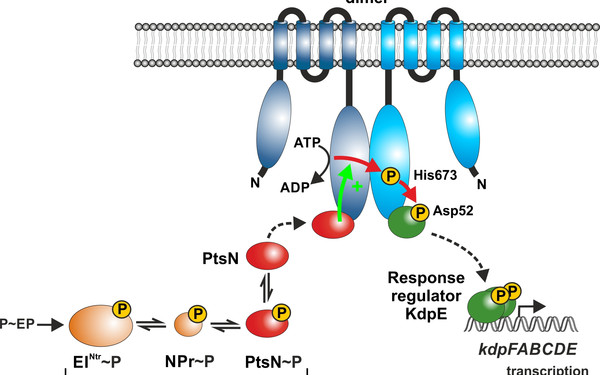
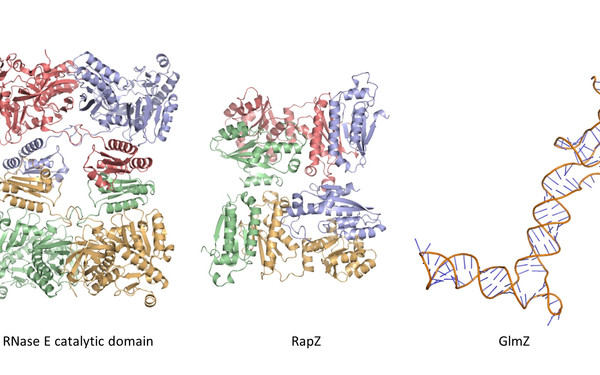
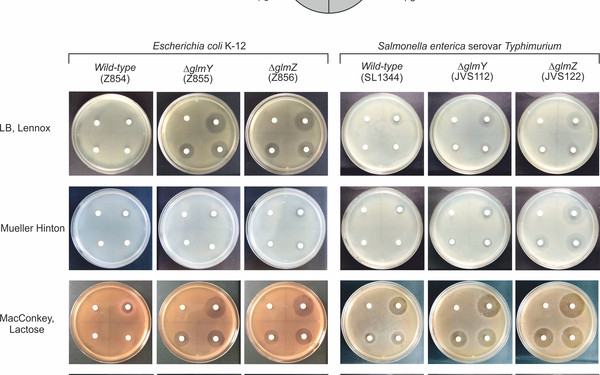
Bacteria live under ever-changing environmental conditions. To survive and propagate, bacteria sense environmental cues and respond to them quickly allowing for adaptation. Our research aims to unravel novel principles and pathways underlying signal-perception, -transduction and cellular regulation in the model organism Escherichia coli and related bacteria including pathogens. We focus on mechanisms acting at the post-transcriptional level including small RNAs and RNA-binding proteins. Moreover, we study two-component and phosphotransferase systems, which monitor the environment and transduce information into the cell. Specifically, we investigate a regulatory network, which controls cell envelope biosynthesis and repair in response to metabolic signals and extracytoplasmic stress. This network involves the trans-envelope signaling complex QseEGF, which employs a regulatory cascade composed of small RNAs GlmY/GlmZ and the RNA binding protein RapZ to achieve envelope homeostasis.