Spotlights
Marco named 'highly cited researcher'
For the fourth time in a row, Marco has been highlighted as highly cited researcher by Clarivate; one among ~60 highly cited scientists in Austria.
TV interview on viruses and synthetic biology
Marco Hein talks with 'Spontan gefragt' host Prof. Markus Hengstschläger and comedian Dr. Omar Sarsam [Interview in German].
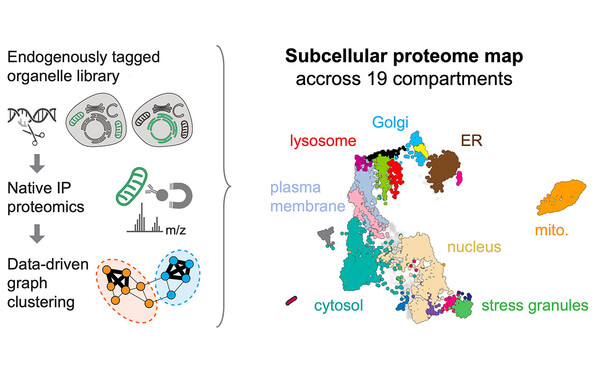
Global organelle profiling reveals subcellular localization and remodeling at proteome scale
With Manu Leonetti's group at the Chan Zuckerberg Biohub, we used immunoprecipitation of intact organelles to map the subcellular localization of the majority of proteins in human cells, and the dynamics of the subcellular proteome upon viral infection with HCoV-OC43.
Browse the resource at organelles.czbiohub.org and read our paper (Hein et al., Cell, 2025)
The Vienna Science and Technology Fund (WWTF) funded our proposal "Deconstructing and Reconstructing Anelloviruses".
Our lab, together with the group of Irene Görzer at the MedUni Center for Virology, will receive 900,000 Euros for four years.
Anelloviruses are ubiquitous viruses that infect virtuall all humans, apparently without causing disease. We're eager to shed light on the almost entirely unknown cell biology of these viruses.
Read more about the proposal and the other projects funded as part of the "Life Sciences 2024 - Synthetic Biology" call.
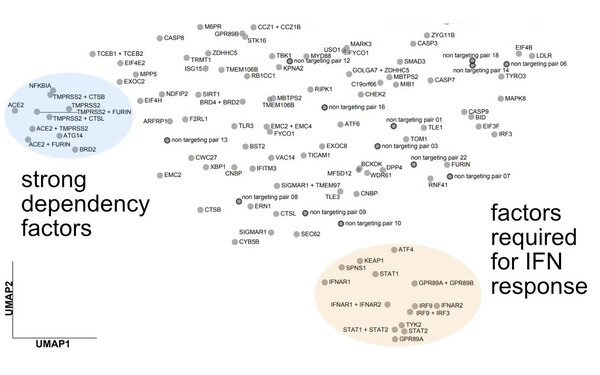
Systematic characterization of SARS-CoV-2 host factors
We used Perturb-seq to map the phenotypic landscapes of host factors of SARS-CoV-2, characterizing what happens to the course of infection when host factors are inactivated. Two classes of factors stood out: factors involved in entry or early events, and those mediating the interferon response (Sunshine et al., Nature Communications, 2023)
Learning from viruses
Interview with Marco Hein about the excitement of discovering something new and what can be learned from viruses about cells and from viral pandemics about science.
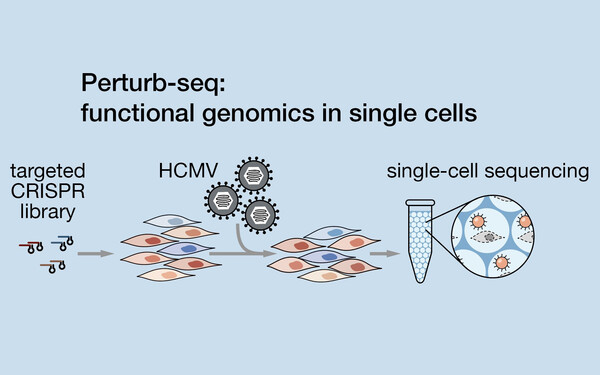
Functional single-cell genomics of HCMV infection
We studied the infection of cytomegalovirus in human cells by a novel, functional single-cell genomics approach (Hein & Weissman, Nature Biotechnology, 2021). Our results reveal a dichotomy between the roles of host and viral factors: viral factors define the trajectory of infection and host factors create the environment permitting the execution of that program.
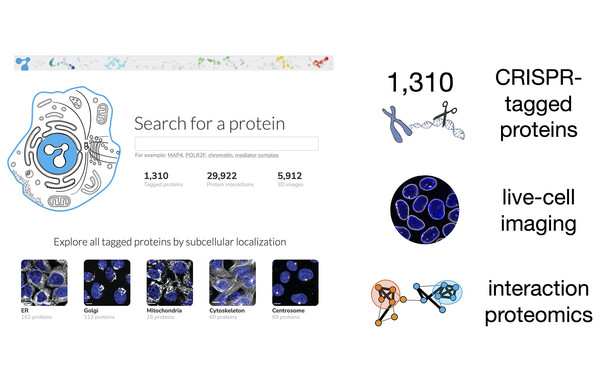
OpenCell: Cartography of human cellular organization
Proud to be part of team OpenCell! Combining endogenous protein tagging, live-cell microscopy and interaction proteomics, we studied the physical and functional organization of the human proteome. Browse the resource at opencell.czbiohub.org and read the paper (Cho et al., Science, 2022)