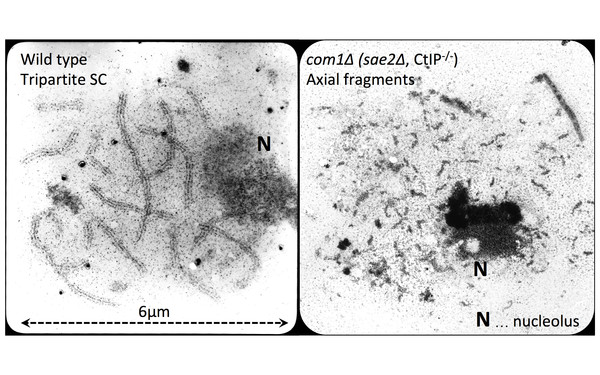
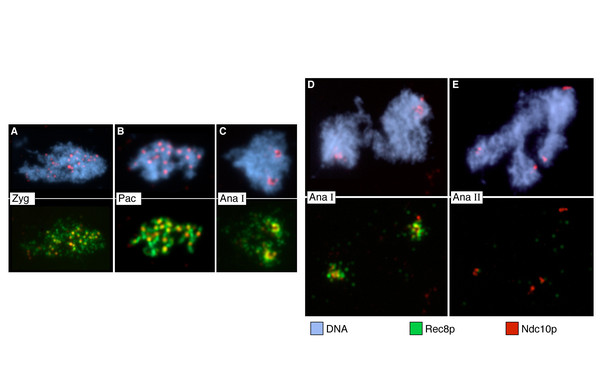
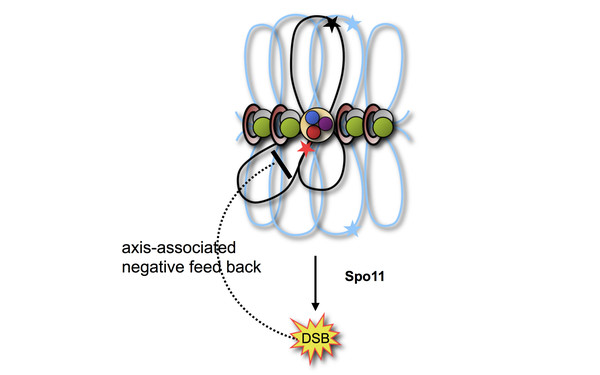
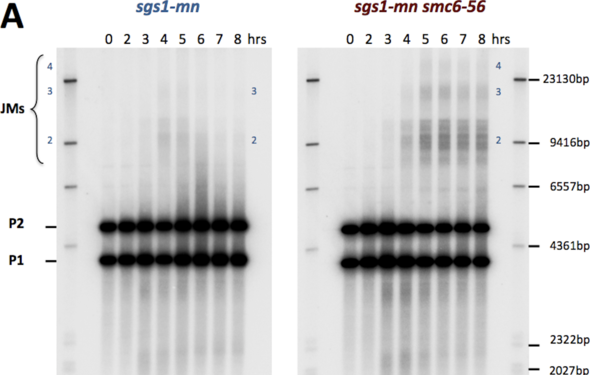
The Question
Chromosomes are the biologically relevant form of organization of the genome in all known forms of life. Eukaryotes, but not prokaryotes, possess basic proteins, which organize DNA as chromatin, possibly to suppress transcriptional noise and chemical instability and to foster phase separation. Eukaryotes also developed sustainable sexual reproduction, involving regular fusion and fission of genomes, the latter known as meiosis, which converts a diploid precursor cell into four haploid products. In meiosis, homologs from each parent must be identified, linked by chiasmata and finally be transported to opposite meiotic products. We currently address the mechanism of identification, initiated by Spo11 mediated double-strand break formation, as well, as homology search and repair. In most organisms a subset of DSBs is turned into crossing over, the basis of the physical linkage between the homologues. Taken together, we study chromosome segregation mediated by meiotic recombination.