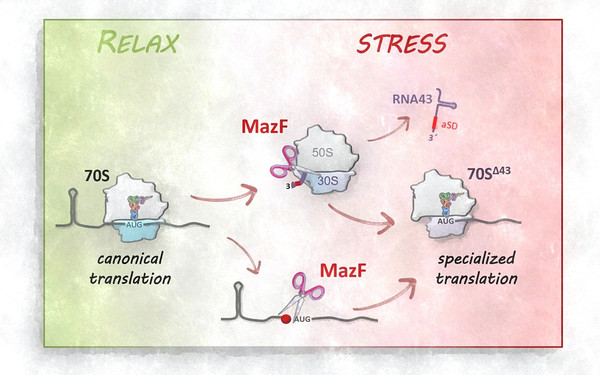
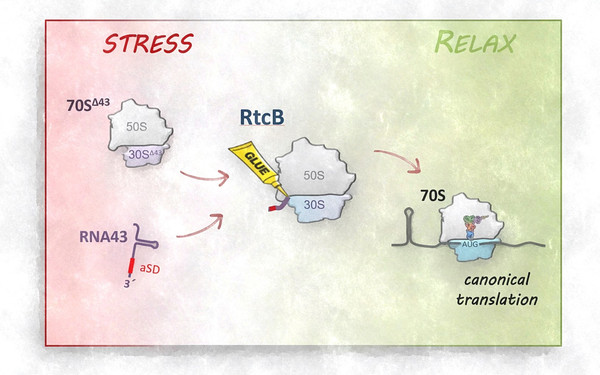
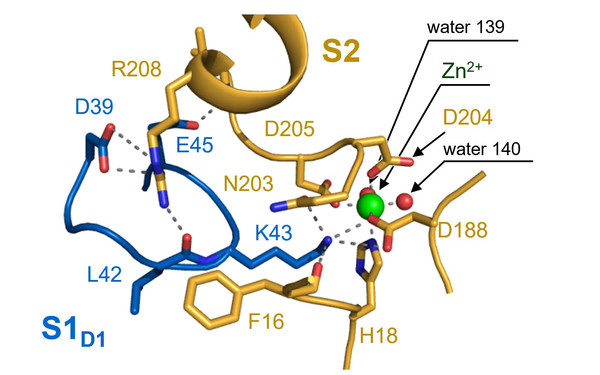
The Question
One of the most intricate and fundamental processes of life is the translation of the genetic code into proteins. Decoding mRNA-based information into the corresponding sequence of amino acids is performed by a complex ribonucleoprotein particle, the ribosome. Traditionally, the ribosome is viewed as a highly conserved machinery with an invariable RNA and protein complement. This perception of invariance has been perpetuated by the determination of high-resolution structures, which implied that ribosomes are homogeneous entities that are not considered to have an intrinsic regulatory capacity. In my group, we aim to challenge this assumption and to decipher mechanisms in bacteria that modulate the translational program by ribosome heterogeneity. In particular, we focus on modifications of ribosomal RNAs and proteins that alter ribosome specificity in response to environmental stress.