Spotlights
Mapping the perturbome network of cellular perturbations
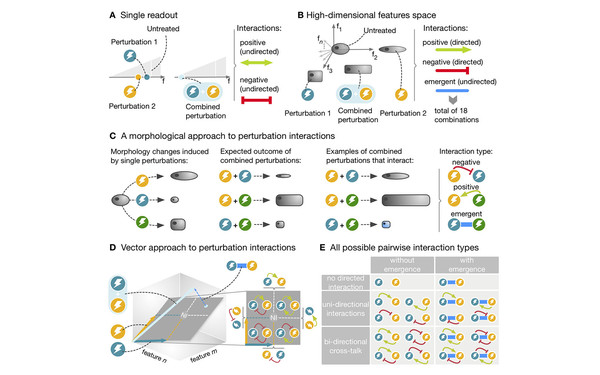
We developed a novel mathematical framework that provides a deeper understanding of how drugs interact with each other. The framework offers the first rigorous approach to quantifying how perturbations with high-dimensional effects influence each other. Our analysis of over 30k drug pairs applied to cell lines identified almost 2000 interactions and sheds new light on how drugs perturb the molecular network within the cell.
[(2019) Caldera et al., Nature Communications. DOI: 10.1038/s41467-019-13058-9]
Using Virtual Reality to dive into molecular networks
Virtual Reality (VR) technology opens up completely new ways of interacting with large, complex data in an intuitive and immersive fashion. We are developing a VR platform for exploring genome-scale molecular networks and identifying genetic mutations that cause rare hereditary diseases. Our platform represents the first application of this technology for state-of-the-art biomedical data analysis.
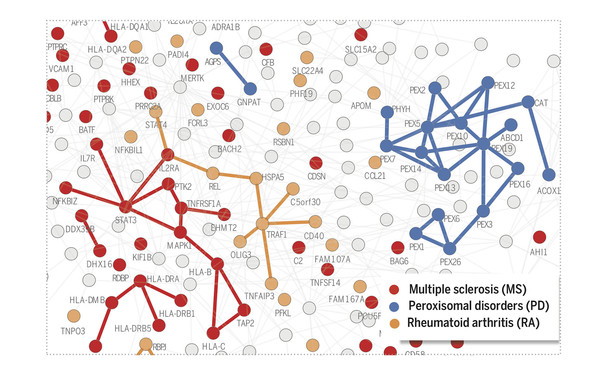
Uncovering relationships between diseases through molecular interactions
Our work laid out the theoretical basis for how molecular networks can be understood as maps to study molecular disease mechanisms. The methodology can be applied to address numerous questions at the forefront of network medicine, from elucidating the molecular origins of relationships between diseases, to interpreting genome-wide association study data or identifying promising drug targets.
[(2015) Menche et al., Science. DOI: 10.1126/science.1257601]