The Question
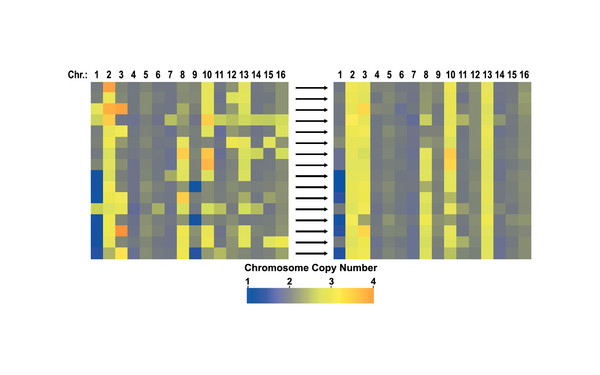
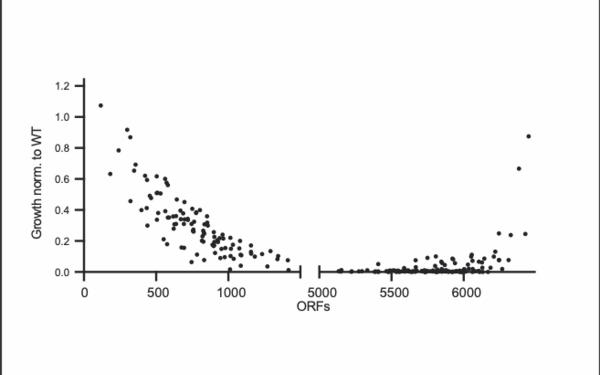
During cell division, errors made during the distribution of chromosomes to the daughter cells result in cells with an abnormal number of chromosomes, which is called aneuploidy. Aneuploidy is the cause of the majority of miscarriages and is also present in ~90 percent of solid tumors. Paradoxically, aneuploidy is universally harmful to cellular fitness, but can be advantageous to the cells under selective conditions. In the Campbell lab, we study how specific patterns of aneuploid chromosomes are selected for over time and how aneuploidy affects cellular physiology and drug resistance.