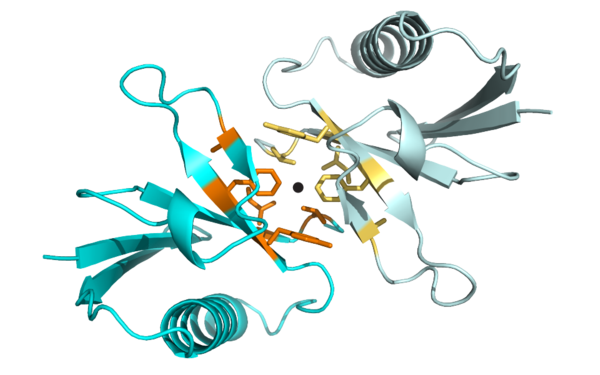
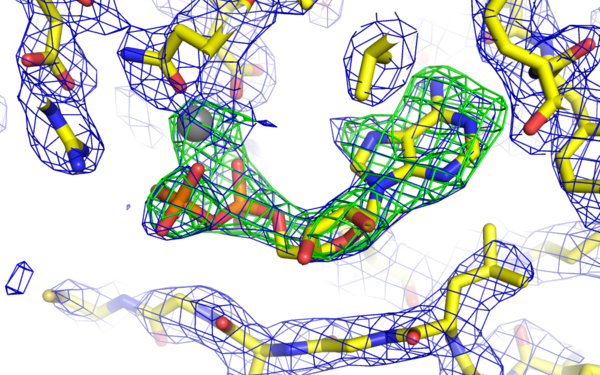
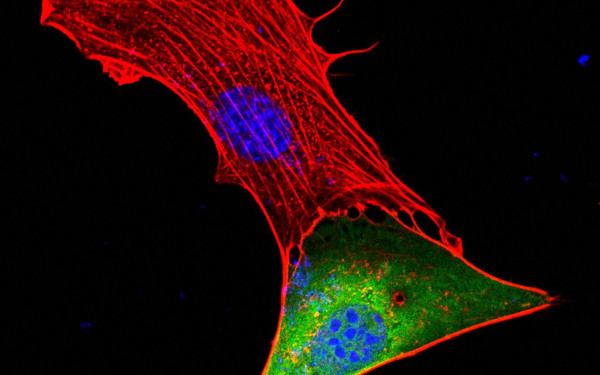
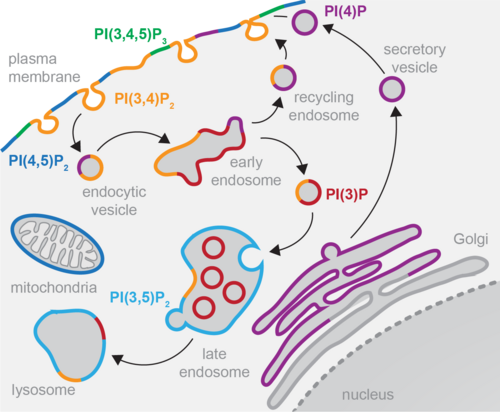
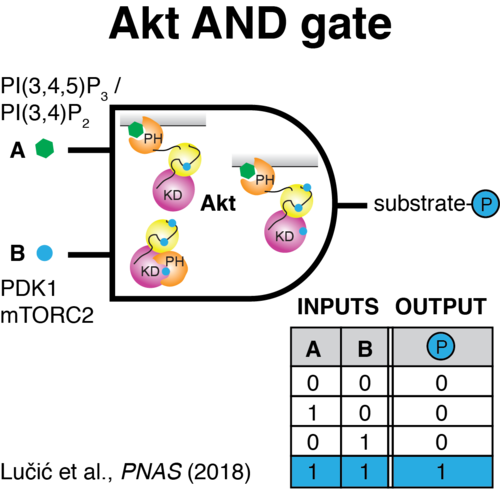
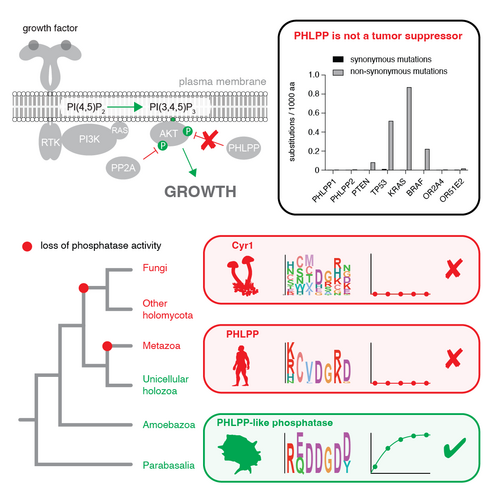
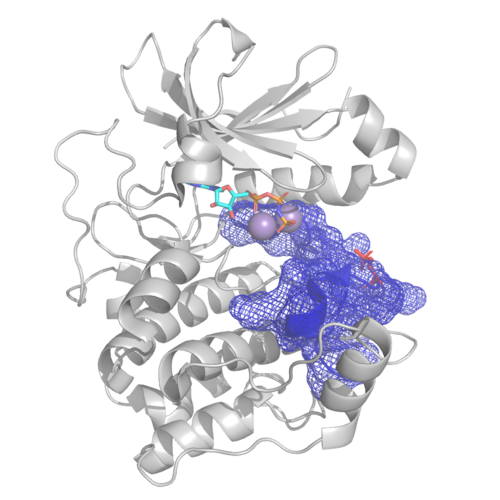
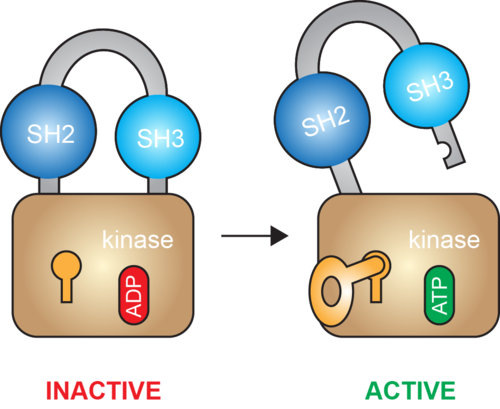
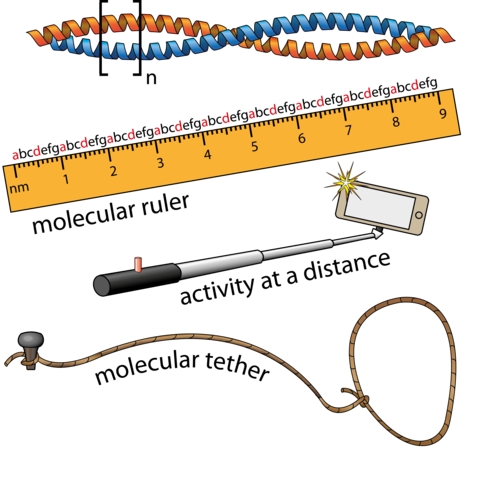
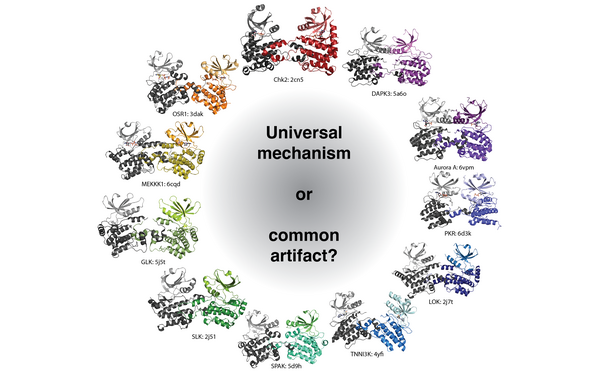
PHLPP2 is a pseudophosphatase that lost activity in the metazoan ancestor.
2025 Proceedings of the National Academy of Sciences of the United States of America(14)
PMID: 40168118
Husremović Tarik, Meier Vanessa, Piëch Lucas, Siess Katharina M, Antonioli Sumire, Grishkovskaya Irina, Kircheva Nikoleta, Angelova Silvia E, Wenzl Karoline, Brandstätter Andreas, Veis Jiri, Miočić-Stošić Fran, Anrather Dorothea, Hartl Markus, Truebestein Linda, Cerron-Alvan Luis M, Leeb Martin, Žagrović Bojan, Hann Stephan, Bock Christoph, Ogris Egon, Dudev Todor, Irwin Nicholas A T, Haselbach David, Leonard Thomas A
Structure and regulation of the myotonic dystrophy kinase-related Cdc42-binding kinase.
2023 Structure (London, England : 1993);31(4):435, 446.e4, 435-446.e4.
PMID: 36854301
Truebestein Linda, Antonioli Sumire, Waltenberger Elisabeth, Gehin Charlotte, Gavin Anne-Claude, Leonard Thomas A
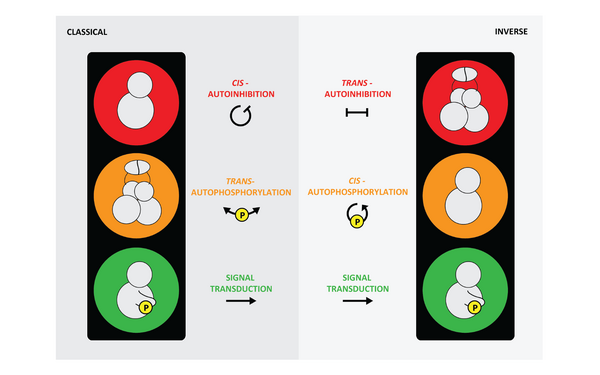
PKD autoinhibition in trans regulates activation loop autophosphorylation in cis
2023 Proceedings of the National Academy of Sciences of the United States of America;120(7):e2212909120.
PMID: 36745811
Reinhardt Ronja, Hirzel Kai, Link Gisela, Eisler Stephan A, Hägele Tanja, Parson Matthew A H, Burke John E, Hausser Angelika, Leonard Thomas A
Activation of the essential kinase PDK1 by phosphoinositide-driven trans-autophosphorylation.
2022 Nature communications;13(1):1874.
PMID: 35387990
Levina Aleksandra, Fleming Kaelin D, Burke John E, Leonard Thomas A
Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation
2021 Proceedings of the National Academy of Sciences of the United States of America;118(33)
PMID: 34385319
Truebestein Linda, Hornegger Harald, Anrather Dorothea, Hartl Markus, Fleming Kaelin D, Stariha Jordan T B, Pardon Els, Steyaert Jan, Burke John E, Leonard Thomas A
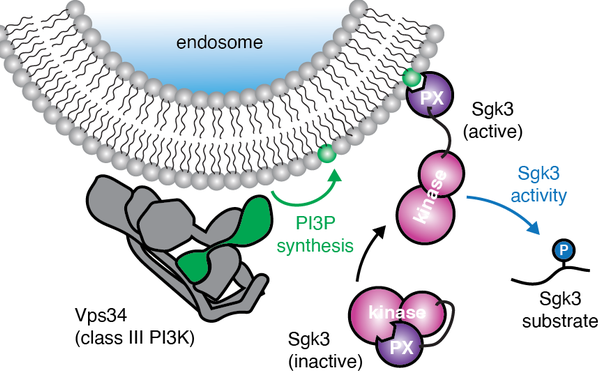
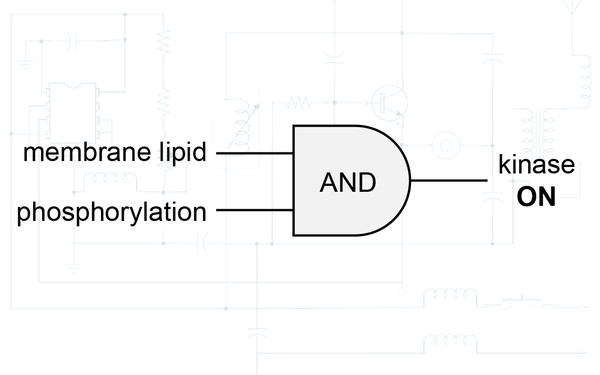
In vitro reconstitution of Sgk3 activation by phosphatidylinositol 3-phosphate.
2021 The Journal of biological chemistry;297(2):100919.
PMID: 34181950
Pokorny Daniel, Truebestein Linda, Fleming Kaelin D, Burke John E, Leonard Thomas A
A ubiquitin-like domain controls protein kinase D dimerization and activation by trans-autophosphorylation.
2019 The Journal of biological chemistry;294(39):14422, 14441, 14422-14441.
PMID: 31406020
Elsner Daniel J, Siess Katharina M, Gossenreiter Thomas, Hartl Markus, Leonard Thomas A
Conformational sampling of membranes by Akt controls its activation and inactivation.
2018 Proceedings of the National Academy of Sciences of the United States of America;115(17):E3940, E3949, E3940-E3949.
PMID: 29632185
Lučić Iva, Rathinaswamy Manoj K, Truebestein Linda, Hamelin David J, Burke John E, Leonard Thomas A
PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes
2017 Molecular cell;65(3):416, 431.e6, 416-431.e6.
PMID: 28157504
Ebner Michael, Lučić Iva, Leonard Thomas A, Yudushkin Ivan
A molecular ruler regulates cytoskeletal remodelling by the Rho kinases.
2015 Nature communications;6:10029.
PMID: 26620183
Truebestein Linda, Elsner Daniel J, Fuchs Elisabeth, Leonard Thomas A