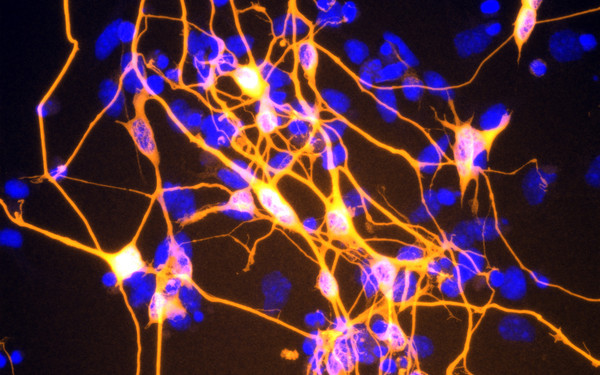
PHF3 regulates neuronal gene expression through the Pol II CTD reader domain SPOC.
2021 Nature communications;12(1):6078.
PMID: 34667177
Appel Lisa-Marie, Franke Vedran, Bruno Melania, Grishkovskaya Irina, Kasiliauskaite Aiste, Kaufmann Tanja, Schoeberl Ursula E, Puchinger Martin G, Kostrhon Sebastian, Ebenwaldner Carmen, Sebesta Marek, Beltzung Etienne, Mechtler Karl, Lin Gen, Vlasova Anna, Leeb Martin, Pavri Rushad, Stark Alexander, Akalin Altuna, Stefl Richard, Bernecky Carrie, Djinovic-Carugo Kristina, Slade Dea
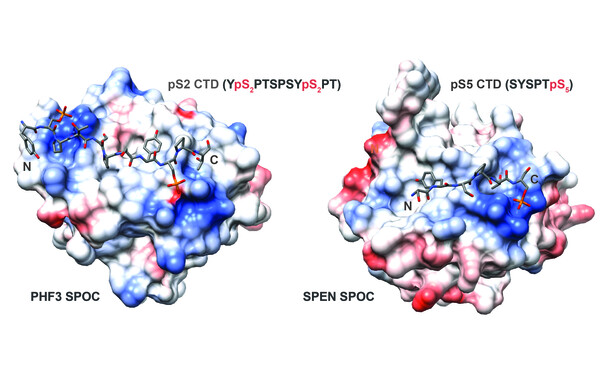
The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators.
2023 Nature communications;14(1):166.
PMID: 36631525
Appel Lisa-Marie, Franke Vedran, Benedum Johannes, Grishkovskaya Irina, Strobl Xué, Polyansky Anton, Ammann Gregor, Platzer Sebastian, Neudolt Andrea, Wunder Anna, Walch Lena, Kaiser Stefanie, Zagrovic Bojan, Djinovic-Carugo Kristina, Akalin Altuna, Slade Dea
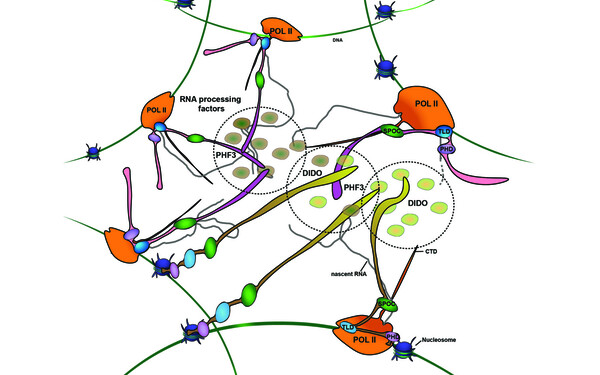
The SPOC proteins DIDO3 and PHF3 co-regulate gene expression and neuronal differentiation.
2023 Nature communications;14(1):7912.
PMID: 38036524
Benedum Johannes, Franke Vedran, Appel Lisa-Marie, Walch Lena, Bruno Melania, Schneeweiss Rebecca, Gruber Juliane, Oberndorfer Helena, Frank Emma, Strobl Xué, Polyansky Anton, Zagrovic Bojan, Akalin Altuna, Slade Dea
SPOC domain proteins in health and disease.
2023 Genes & development;37(5-6):140, 170, 140-170.
PMID: 36927757
Appel Lisa-Marie, Benedum Johannes, Engl Magdalena, Platzer Sebastian, Schleiffer Alexander, Strobl Xué, Slade Dea
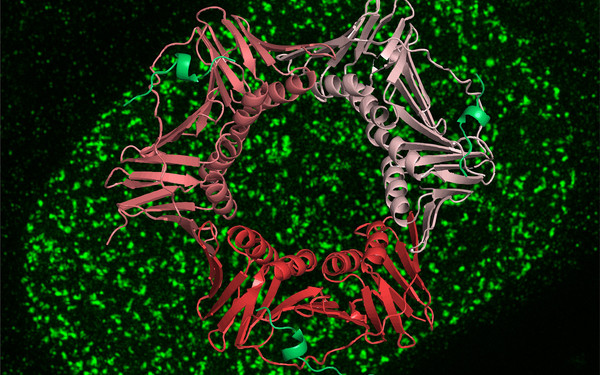
A novel non-canonical PIP-box mediates PARG interaction with PCNA.
2017 Nucleic acids research;45(16):9741, 9759, 9741-9759.
PMID: 28934471
Kaufmann Tanja, Grishkovskaya Irina, Polyansky Anton A, Kostrhon Sebastian, Kukolj Eva, Olek Karin M, Herbert Sebastien, Beltzung Etienne, Mechtler Karl, Peterbauer Thomas, Gotzmann Josef, Zhang Lijuan, Hartl Markus, Zagrovic Bojan, Elsayad Kareem, Djinovic-Carugo Kristina, Slade Dea
PARP inhibition causes premature loss of cohesion in cancer cells.
2017 Oncotarget;8(61):103931, 103951, 103931-103951.
PMID: 29262611
Kukolj Eva, Kaufmann Tanja, Dick Amalie E, Zeillinger Robert, Gerlich Daniel W, Slade Dea
PARP and PARG inhibitors in cancer treatment.
2020 Genes & development;34(5-6):360, 394, 360-394.
PMID: 32029455
Slade Dea
SIRT2 regulates nuclear envelope reassembly through ANKLE2 deacetylation.
2016 Journal of cell science;129(24):4607, 4621, 4607-4621.
PMID: 27875273
Kaufmann Tanja, Kukolj Eva, Brachner Andreas, Beltzung Etienne, Bruno Melania, Kostrhon Sebastian, Opravil Susanne, Hudecz Otto, Mechtler Karl, Warren Graham, Slade Dea
Direct measurement of protein-protein interactions by FLIM-FRET at UV laser-induced DNA damage sites in living cells.
2020 Nucleic acids research;48(21):e122.
PMID: 33053171
Kaufmann Tanja, Herbert Sébastien, Hackl Benjamin, Besold Johanna Maria, Schramek Christopher, Gotzmann Josef, Elsayad Kareem, Slade Dea