National Scientific Research Fund (FWF) Standalone Project
Project title: "NMR investigations of the hyperphosphorylated IDP Osteopontin" (P28359)
National Scientific Research Fund (FWF) Standalone Project
Project title: “Structural Dynamics of IDPs probed by Cross-Correlated NMR Spin Relaxation" (P 28359)
WWTF Vienna Science and Technology Fund
LS17-008, Structure Zoom (as Co-PI with Christian Becker)
EU - FP7
ITN: FLUOR21 participant
Doctoral Program "Integrative Structural Biology"
2016-2019: The Group Konrat participates in the special Doctoral Program "Integrative Structural Biology" reviewed and funded by the Austrian Science Fund FWF.
Research Partners (companies):
Novartis, Boehringer-Ingelheim, UCB, Alkahest, IDPharma, Everpharma, Neuropore
Christian Doppler Laboratory for High-Content Structural Biology and Biotechnology
Project-leader: Robert Konrat
Business partner: Boehringer Ingelheim RCV GmbH & Co KG
Duration: 01.02.2017 - 31.01.2025
CD Laboratory for knowledge-based structural biology and biotechnology
Christian Doppler Research Association
University of Vienna
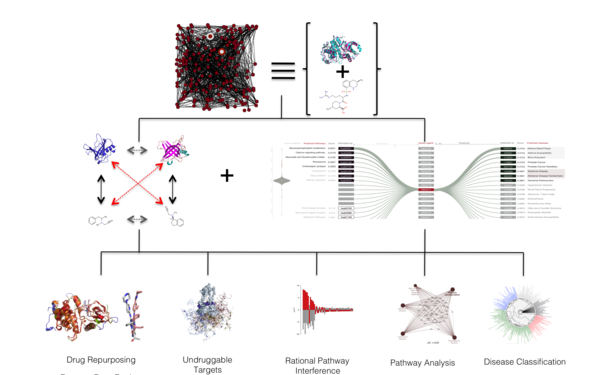
Description: The scientists will primarily investigate protein structure, which is crucial for the development of new therapeutic methods for a great variety of diseases ranging from Alzheimer’s and infectious diseases to cancer. This renders it a most valuable tool in biomedical research. In order to gain new insights into protein structure, function and interactions that could potentially be translated into new therapies, the researchers will employ a combination of bioinformatics, protein production and high-end biophysical and structural biology techniques. The collected information will ultimately be combined in an information pipeline for both research and biotechnology.
National Scientific Research Fund (FWF) Standalone Project
Project title: "NMR Spin Relaxation to probe Side-Chain Dynamics in IDPs" (P35098)
Member of Doctoral Program "Liquid-liquid Phase Separation in Biology – Ellipse"
Liquid-liquid Phase Separation in Biology – Ellipse, funded by the Austrian Science Fund FWF.
2. Platz Houska Preis 2023
Project title: Computergestützte Strukturbiologie